Around 18 months ago we began developing a platform to provide a collaborative environment for running GWAS using the UKBiobank data. To date we have completed 20,000 GWAS on automated phenotypes and 400 GWAS on curated phenotypes. These are being made available via the MR-Base platform with regular updates and an anticipated full release date of December 2018.
Little bit of background
The main aims of the platform were firstly to create an easy to use single job submission/monitoring sheet, enabling researchers to perform complex and computationally demanding analysis without having to be experienced with a HPC environment. Secondly, to provide a uniform approach to a complex method, which is of particular importance for downstream meta-analysis. Thirdly to run GWAS on all suitable UKBiobank traits in accordance with this UKBiobank project proposal. This post does not really cover the first two aspects, details of which can be found here and here. It is more an overview of the pipeline, the semi-automated GWAS of all traits and the issues we encountered along the way.
Phesant phenotypes and HPC
For the large scale automated GWAS analysis, the first step was to transform the UKBiobank phenotype data into a format that could be handled programmatically. For this we used the wonderful Phesant tool, creating > 20,000 traits in one of four formats, continuous, binary, categorical ordered and categorical unordered, the latter of which was ignored. This left 20,812 traits to be batch processed through the GWAS pipeline in a semi-automated manner, i.e. with minimal hand-holding. Periodically batches of 1,000 GWAS were added to the sheet, whilst allowing custom phenotypes to be inserted above a batch, running with preference over the batched jobs.
We are aware that there are far more efficient ways to run GWAS on 20,000 traits, e.g. Ben Neale’s GWAS project using Hail. However, we decided to stick with BOLT-LMM for reasons discussed here, running one trait per node. Many thanks to Po-Ru Loh for his help with all our questions, especially pointing out that we didn’t need to split by chromosome! And, thanks to some well timed purchasing we had 44 dedicated nodes each with 28 cores, so could perform around 200 GWAS per day. The semi-automated GWAS have been running in batches over the last year, allowing time on the nodes for custom traits and other projects.
Can a Google Sheet act as a front and backend?
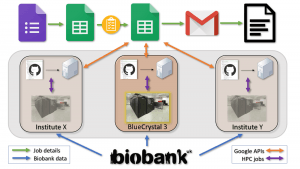
This project seemed like a good time to test how feasible it was to control an application using a Google Sheet. On the whole, it has worked quite well. Would I do it again, probably not. Initially we aimed to provide a pipeline that could run at multiple locations, with a single Google Sheet controlling everything. Users would be able to see all jobs (to avoid duplication), submit their own jobs, and behind the scenes machines at various institutes would identify jobs marked for them, and seamlessly send the jobs off to the HPC of the specified institute. To date, the pipeline has run in Bristol only, so it became a slightly easier task to manage. We did however migrate from BlueCrystal3 (in Bristol with shared filesystem mounted), to BlueCrystal4 (in Slough without said filesystem), so in a sense we did set it up in two locations 🙂
The image at the top displays the main steps in the pipeline. An entry is made in a Google Form or Sheet specifying the details of a particular GWAS. A virtual machine connected to BlueCrystal4 monitors the sheet using the Google Sheet API. When a new job is found and passes various QC checks the BOLT-LMM submission script is passed to the cluster, and the job submitted. The VM continually checks the output and when complete, the data are cleaned up, and the user is alerted via email. In practice that worked, except for the hitting API write quote limit and reaching the cell limit of a Google Sheet. Not being aware of this limit we were surprised to see a non-responding grey screen where all our GWAS data had been. Fortunately, after very slowly and carefully removing all completed jobs the Sheet became active again. This did however, impact the usefulness of the front end as now it wasn’t possible to see the completed traits.
Output and usage
To date we have completed over 20,000 GWAS, with 6,000 added to MR-Base so far. These are being used by researchers all over the world primarily via our MR-Base web application, R-package and API. The infrastructure behind all that will be described in detail at a later date. Suffice to say we are currently managing millions of API calls per week from hundreds of users.
This work has involved many members of the MRC-IEU, including Ruth Mitchell, Gibran Hemani, Lavinia Paternoster, Tom Gaunt, Tom Dudding and myself.

Have you all published descriptions/ definitions of the Phesant generated traits for GWAS uploaded to the MR Base library? Thanks!
Hi Daniel,
Not yet. Currently you just have to match the trait name to the UKBiobank showcase – http://biobank.ctsu.ox.ac.uk/crystal/search.cgi. We’re hoping to have something more official up and running soon.